BESPOKE BIOINFORMATICS PIPELINE DESIGN
UNRAVELING HIGH COMPLEXITY RNA-SEQ DATA
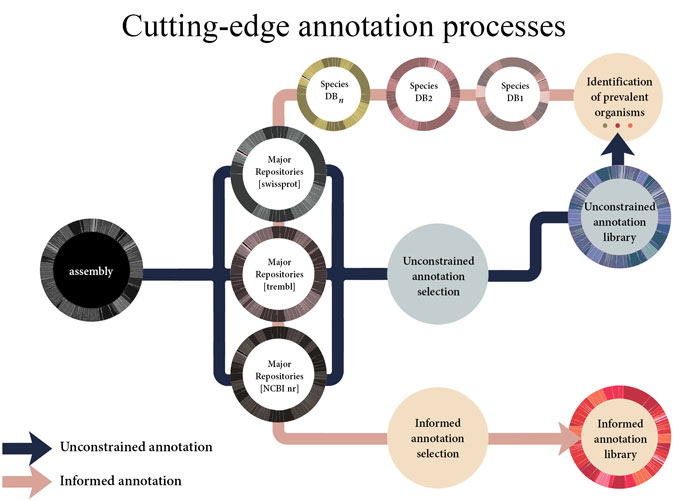
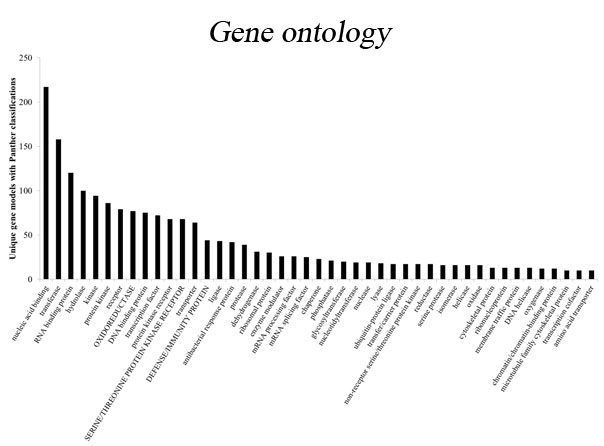
COMPREHENSIVE MULTI-SPECIES ANNOTATIONS
EXPLOIT THE ENTIRETY OF RICH NEXT-GENERATION SEQUENCING DATA
USER FOCUSED OUTPUT
FROM ROBUST BIOINFORMATICS SOLUTIONS TO FULL GENETICS INTERPRETATION
Transcriptomic
Solutions
Transcriptomics
Solutions
SOLUTIONS
QUALITY
ACCESSIBILITY
Thorough pipelines to overcome any massive Next-Generation Sequencing complexity
High resolution bioinformatics, robust data integrity, cutting-edge biological interpretation
Comprehensive,
user-tailored output
ABOUT TRANSCRIPTOMICS SOLUTIONS
Analyzing next-generation sequencing data requires the confident selection of effective bioinformatic approaches driven by a modern understanding of biology. We believe that through tight collaboration between the client and our multi-disciplinary team, the best pipeline for any data set can be designed. Central to Transcriptomic Solutions’ approach is the rejection of any black box within bioinformatics pipelines as next-generation sequencing data sets are both highly complex and unique. Towards this end, the collective expertise of the team will be brought to bear on any data interpretation issues, delivering solutions through robust integration of modern systems biology and cutting-edge computational science.
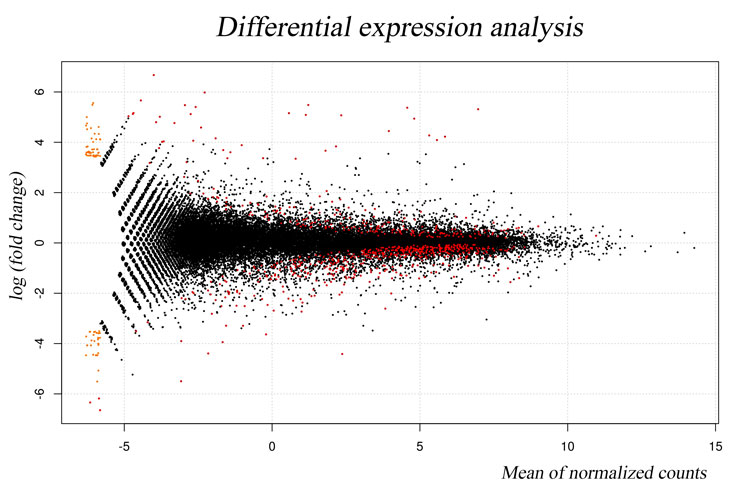
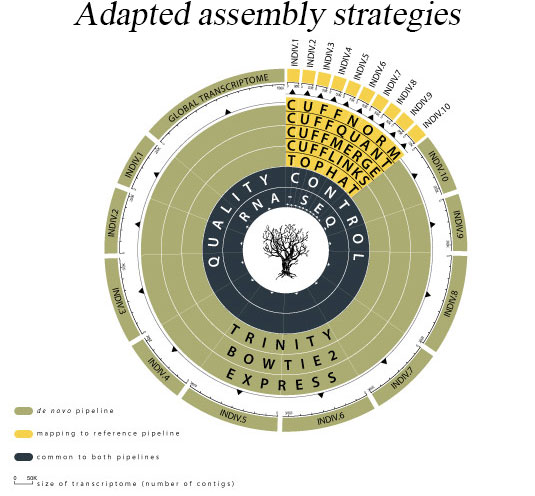
COMPLEXITY MATTERS
We specialize in non-model organism, high uncertainty and high complexity data, including the very cutting-edge of metaorganismal systems. Additionally, we can produce rich data output from all well characterized model organisms, designed for ease of interpretation and publication by biologists. Although less controlled or environmental experiments can be attractive in terms of application driven biological impact, they generally demand very high computer power and advanced computing skills that may be challenging and time consuming for the majority of research scientists. Transcriptomics Solutions provides a fast and inexpensive option to explore such data to the highest possible standard of quality without the need for a reduction of complexity.
CONSULTING SERVICES OFFERED
EXPERIMENTAL DESIGN
LABORATORY PROTOCOLS
HARDWARE DESIGN
NGS DATA PIPELINE
SCRIPT DEVELOPMENT
ACCESSIBILITY TAILORING
BIOLOGICAL ANALYSIS SUPPORT

DR. EMMANUEL GONZALEZ
Bioinformatician, Computational Chemist
Norwegian Center of Excellence (CTCC)
Plant Biology Institute of Montreal
MEET THE TEAM
WHAT WE CAN DO FOR YOU
We have a SOLUTION
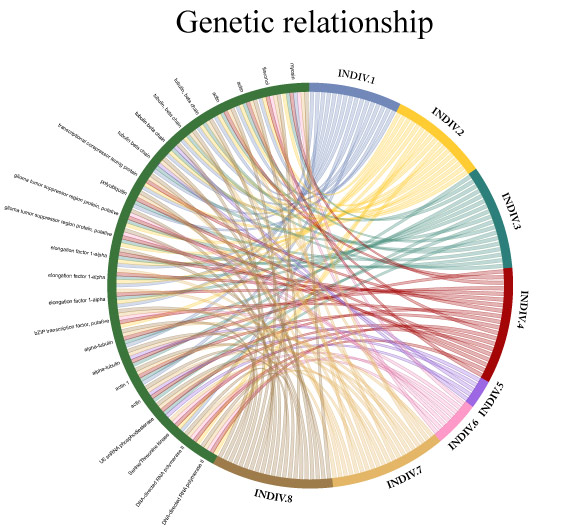
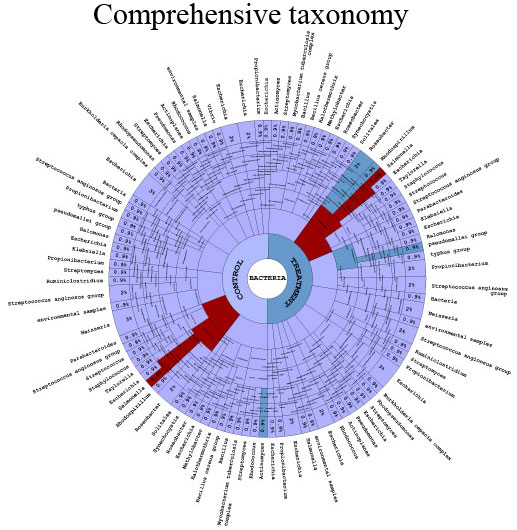

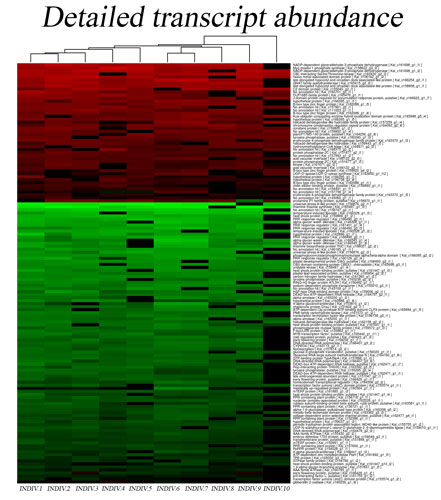
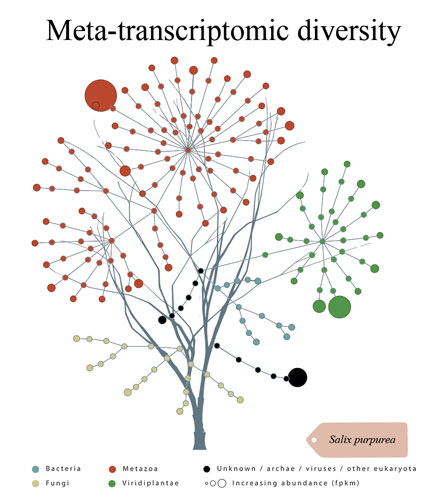

DR. NICHOLAS BRERETON
Biologist, Meta-Organism Geneticist
Imperial College in London
Plant Biology Institute of Montreal
Give us, if possible, the sequencing platform used :
ABI SOLID, HELICOS, ILLUMINA, ION TORRENT, LS 454, OXFORD NANOPORE, PACBIO